### Kaggle Submission Code
"""
//kaggle submission
//Biological Response
--> random forest classifier
Author: Robertmitchellv
Date: Dec 16, 2104
Revised: Dec 22, 2014
"""
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
def main():
# create the training + test sets
try:
data = pd.read_csv('Data/train.csv')
except IOError:
print("io ERROR-->Could not locate file.")
target = data.Activity.values
train = data.drop('Activity', axis = 1).values
test = pd.read_csv('Data/test.csv').values
# create and train the random forest and call it 'rf'
# --> n_estimators = the number of trees in this forest, viz.
# 100 trees of forest
# --> n_jobs set to -1 will use the number of cores present on your system.
rf = RandomForestClassifier(n_estimators = 100, n_jobs = -1)
# fit(X, y[, sample_weight]) = build a forest of tress from the
# training set (X, y)
rf.fit(train, target)
# predict_proba(X) predict class probabilities for X as list
predicted_probs = [x[1] for x in rf.predict_proba(test)]
# prep data for use in pd.Series
molID, predictProbs = prepData(predicted_probs)
# use a dictionary with keys as col headers and values as lists pulled from
# previous prep function
df = {'MoleculeID': molID, 'PredictedProbability': predictProbs}
# pandas DataFrame = a tabular datastructure like a SQL table
predicted_probs = pd.DataFrame(df)
# write predicted_probs to file with pandas method .to_csv()--add header
# for submission
try:
predicted_probs.to_csv('Data/submission.csv', index = False)
print("File successfully written; check 'Data' folder")
except IOError:
print("io ERROR-->Could not write data to file.")
# preparing data for conversion to pd.DataFrame
def prepData(alist):
# prepare list to be converted to pandas Series
colOne = []
colTwo = []
idx = 1
# for loop to set MoleculeID to match the benchmark;
# place values into list for easier wrangling as pd.Series
for i in alist:
colOne.append(idx)
colTwo.append(i)
idx += 1
return colOne, colTwo
# call the main function
main()This is a very old post. Please check the sklearn documentation for examples of how to run a Random Forest Classifier. This remains here as a record for myself
I have seen kaggle mentioned on twitter a lot; mostly by the data scientists and researchers I look up to, but there’s never been much confidence that the site was for me in any way—mostly because I was a long way from my dream data science job with yet so much to learn. Notwithstanding, I cannot help but try and hack my way to my destination! I think it’s a part of my learning process: thrust myself in the midst of something I don’t understand, get stuck, try to get unstuck, finish with some understanding of what I was doing.
So, when I saw this post by Chris Clark, I thought that it was about time I try and hack my way from recently learning Python to machine learning with SciKit-Learn&—why not!?&—I thought.
It reminded me of when I decided to sign up with an account at GitHub; I was initially intimidated because it was new to me. Now, I use git in the command line, host my website there, and use it for almost everything (still learning new things about git everyday as well).
Chris’s post was excellent but there was one problem: the code was aimed at Python 2.7 users and I had just spent the previous semester learning Python 3 (which means I don’t really know 2.7; and avoid it all the time “where are the parens for this print statement??”). As a personal challenge, I decided to use the code and update it to Python 3, which was both fun and challenging (I’m measuring ‘update’ to mean, ‘running in my Python 3.4 interpreter without error messages’). This may be an easy task but there were a few snags for me.
In the spirit of trying to document the things I learn, I’ve decided to chronical my results here&—if there are any errors or issues with this code, please let me know so I can try to correct, learn, and grow! I also found Chris’s updated code on GitHub, which uses Pandas and I’ve been trying to get started with Pandas as well so; win, win.
As an aside, I use Anaconda and Vim for the enviornment and editing, respectively. My code can be found on GitHub.
The Submission was a part of the Predicting a Biological Response competition, and the training, test, and benchmark data sets are provided.
Since the competition wants us to predict binary values, Chris notes that this data set is a good introduction to ensemble classifiers, because the prediction is a binary value (0 or 1). It was also great to take a closer look at both the Pandas and SciKit-Learn’s documentation to troubleshoot. I tried to use the comments to explain as much as possible so future me will not be baffled, which I can say is helpful since I’m looking at this one month out and it makes total sense (at least to me).
After performing this–Chris suggested to submit to kaggle; being an extra careful person by nature, I just had to perform the evaluation and cross validation first (I don’t know if any of you feel the same way). Unfortunately, I don’t really understand how the code works–this is one of the problems when hacking through tutorials.
### Evaluation/Logloss
"""
//kaggle submission
//Biological Response
--> evaluation function (from Grunthus' post)
"""
import scipy as sp
def logloss(act, pred):
""" Vectorised computation of logloss """
#cap in official Kaggle implementation,
#per forums/t/1576/r-code-for-logloss
epsilon = 1e-15
pred = sp.maximum(epsilon, pred)
pred = sp.minimum(1-epsilon, pred)
#compute logloss function (vectorised)
ll = sum( act*sp.log(pred) +
sp.subtract(1,act)*sp.log(sp.subtract(1,pred)))
ll = ll * -1.0/len(act)
return llThe cross validation was trickier to understand, which I think is mostly due to my not really understanding what ensemble classifiers do, how the random forest classifier works, and more specifically; what training, test, and target data do within machine learning. This gave chase through the SciKit-Learn documentation and other resources online to get a better understanding of what the code was doing&—there’s a lot to learn! The interesting aspect is how the SciKit-Learn reserves some actual data that it can test against the classifier’s predicted values. I tried to show in the comments how I was understanding what the code did at the time.
### Cross Validation
"""
//kaggle submission
//Biological Response
--> cross validation
"""
from sklearn.ensemble import RandomForestClassifier
from sklearn.cross_validation import KFold
import numpy as np
import pandas as pd
import logloss
def main():
#read data from csv; use nparray to create the training + target sets
try:
train = pd.read_csv('Data/train.csv')
except IOError:
print("io ERROR-->Could not locate file.")
target = np.array([x[0] for x in train])
train = np.array([x[1:] for x in train])
# in this case we'll use a random forest, but this could be any classifier
model = RandomForestClassifier(n_estimators = 100, n_jobs = -1)
# simple K-Fold cross validation. 10 folds.
cv = KFold(n = len(train), n_folds = 10, indices = False)
#iterate through the training and test cross validation segments and
#run the classifier on each one, aggregating the results into a list
results = []
for traincv, testcv in cv:
prob = model.fit(train[traincv], target[traincv]).predict_proba(train[testcv])
results.append(logloss.llfun(target[testcv], [x[1] for x in prob]))
#print out the mean of the cross-validated results
print('Results: ', str(np.array(results).mean()))
# call main function
main()After I was able to execute the submission, logloss, and cross validation code without any errors, I submitted my code to kaggle. It was an exciting moment waiting to see what kind of score I would have recieved had I actually participated in the competition. I would have placed at 325 (well, I would have tied with another user for 325th); check out my results below.
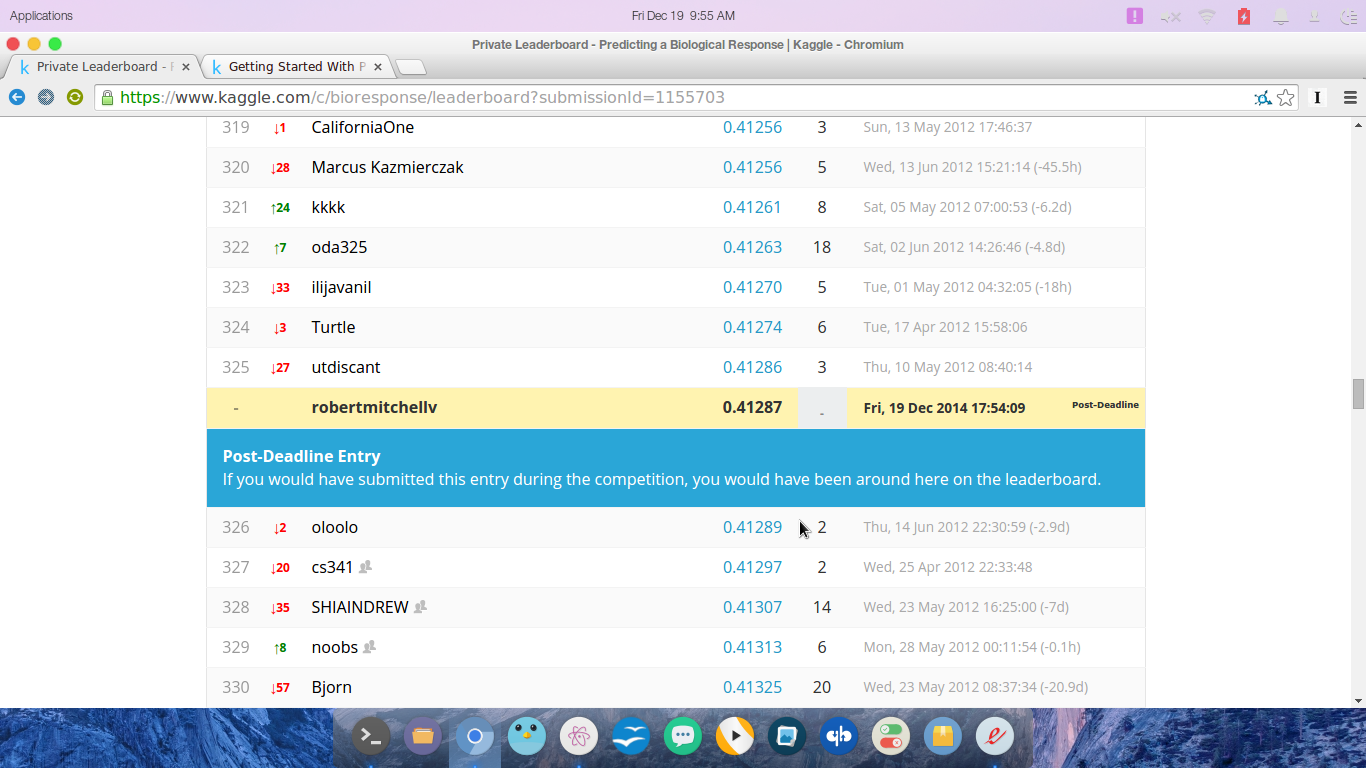
Well, that wraps up my first submission to kaggle. I really hope this is the first of many. Right now I’m working through the Think Stats + Think Bayes books to refresh my stats knowledge. I’m trying to find time to work on the Titanic tutorial through kaggle as well as perhaps throw a hat in the ring for Booz Hamilton’s Data Science Bowl. There’s so much to learn and I can’t wait for these concepts to become more natural and familiar.